Prashant Govindarajan
Hello and வணக்கம்! I am a fourth-year Computer Engineering PhD candidate at Mila-Quebec AI Institute and Polytechnique Montréal (engineering school of UdéM), working under Sarath Chandar. I am keenly interested in reinforcement learning and large language models for scientific discovery and beyond. In collaboration with Intel, I developed offline and online reinforcement learning methods for crystalline material design using first-principles. I also worked with Ansys on LLMs for automating Computer-Aided Design (CAD). Currently, I am working on improving exploration in RL fine-tuning of LLMs. My research was supported by the PBEEE scholarship for international students.
I was previously a dual degree student at the Indian Institute of Technology Madras, where I worked under Balaraman Ravindran and Karthik Raman on target-specific drug design. Besides academics, I like playing chess, frisbee, traveling, and cooking. Lately, I’ve been learning to snowboard and play the flute. I organize social activites and outings in my lab. Check out some cool photos here: https://chandar-lab.github.io/photos/ !
I am always open to discussing research and potential collaborations. Please feel free to reach out 😁!
I am actively seeking research internships in the areas of LLMs, reinforcement learning, and AI for Science.
Publications
- Woo, Kowen, Prashant Govindarajan, and Sarath Chandar. Benchmarking Machine Learning Potentials for Crystal Structure Relaxation. In NeurIPS 2025 AI for Science Workshop.
- Govindarajan, Prashant*, Davide Baldelli*, Jay Pathak, Quentin Fournier, and Sarath Chandar. 2026. “CADmium: Fine-Tuning Code Language Models for Text-Driven Sequential CAD Design.” Transactions on Machine Learning Research.
- Govindarajan, Prashant, Mathieu Reymond, Antoine Clavaud, Mariano Phielipp, Santiago Miret, and Sarath Chandar. CrystalGym: A New Benchmark for Materials Discovery Using Reinforcement Learning. In AI for Accelerated Materials Design-ICLR 2025.
- Govindarajan, Prashant, Mathieu Reymond, Santiago Miret, Mariano Phielipp, and Sarath Chandar. Crystal Design Amidst Noisy DFT Signals: A Reinforcement Learning Approach. In AI for Accelerated Materials Design-NeurIPS 2024.
- Govindarajan, Prashant, Mathieu Reymond, Santiago Miret, Antoine Clavaud, Mariano Phielipp, and Sarath Chandar. A Reinforcement Learning Pipeline for Band Gap-directed Crystal Generation. In AI for Accelerated Materials Design-Vienna 2024.
- Govindarajan, Prashant, Santiago Miret, Jarrid Rector-Brooks, Mariano Phielipp, Janarthanan Rajendran, and Sarath Chandar. Learning Conditional Policies for Crystal Design Using Offline Reinforcement Learning. Digital Discovery (2024).
- Govindarajan, Prashant, Santiago Miret, Jarrid Rector-Brooks, Mariano Phielipp, Janarthanan Rajendran, and Sarath Chandar. Behavioral Cloning for Crystal Design.” In Workshop on Machine Learning for Materials, ICLR 2023.
News
- January 2026 CADmium is now published in TMLR!
- December 2025 Attending NeurIPS 2025 in San Diego and presenting in the AI4Science workshop.
- October 2025 See me at the COLM conference!
- August 2025 Teaching Assistant for INF8245AE, Machine Learning by Sarath Chandar
- July 2025 Released CADmium paper and dataset!
- June 2025 Selected for FRQNT merit scholarship for international students (PBEEE)
- April 2025 CrystalGym: A New Benchmark for Materials Discovery Using Reinforcement Learning selected for a Spotlight at AI4Mat, ICLR 2025!
- December 2024 Crystal Design Amidst Noisy DFT Signals: A Reinforcement Learning Approach accepted at AI4Mat 2024 (NeurIPS 2024)
- July 2024 A Reinforcement Learning Pipeline for Band Gap-directed Crystal Generation accepted at AI4Mat 2024 (Vienna)
- May 2024 Attended Sciencepreneurship Summer School at EPFL, Switzerland, and our team won the first place in the pitch competition!
- April 2024 Passed my quals! I’m a PhD Candidate now!
- March 2024 Organizing MoML Conference (summer edition) for the second time (MoML 2024)!
- January 2024 Learning Conditional Policies for Crystal Design Using Offline Reinforcement Learning accepted for the AI4Mat-2023 Digital Discovery Special Issue!
- December 2023 Presented poster at AI4Mat Workshop at NeurIPS 2023, New Orleans
- November 2023 Volunteer for Graduate Application Assistance Program for Underrepresented Students in AI
- November 2023 Presented poster at MoML 2023, MIT
- August 2023 Teaching assistant for INF8250AE, Reinforcement Learning by Sarath Chandar
- March 2023 Behavioral Cloning for Crystal Design accepted as workshop paper at ML4Materials workshop, ICLR 2023
- February 2023 Organizer of Molecular ML Conference (MoML 2023) happening on May 29, 2023
- August 2022 Started PhD in Computer Engineering at Mila and Polytechnique Montreal, advised by Sarath Chandar
- July 2022 Graduated from IIT Madras with a dual degree in Biological Sciences and Data Science
- June 2022 Defended M.Tech thesis titled “Graph generative models for binding site-specific molecule generation”
- May 2022 “Generating drug-like molecules from gene expression signatures using transformer model” accepted as poster at MLCSB-COSI, ISMB 2022
- April 2022 Presented poster for thesis work at RBCDSAI Annual Research Showcase
- April 2022 Selected for international travel bursary to attend Amii’s AI Week 2022
- January 2020 Selected for the Khorana Program for Scholars to undertake research internship in the USA
Projects
Fine-tuning LLMs for automating Computer-aided Designing
With Ansys, we developed CADmium, a new open-source dataset consisting of natural language descriptions for 170k+ 3D CAD objects. We propose a novel LLM fine-tuning workflow with code LLMs, and augment existing evaluation schemes with new metrics. 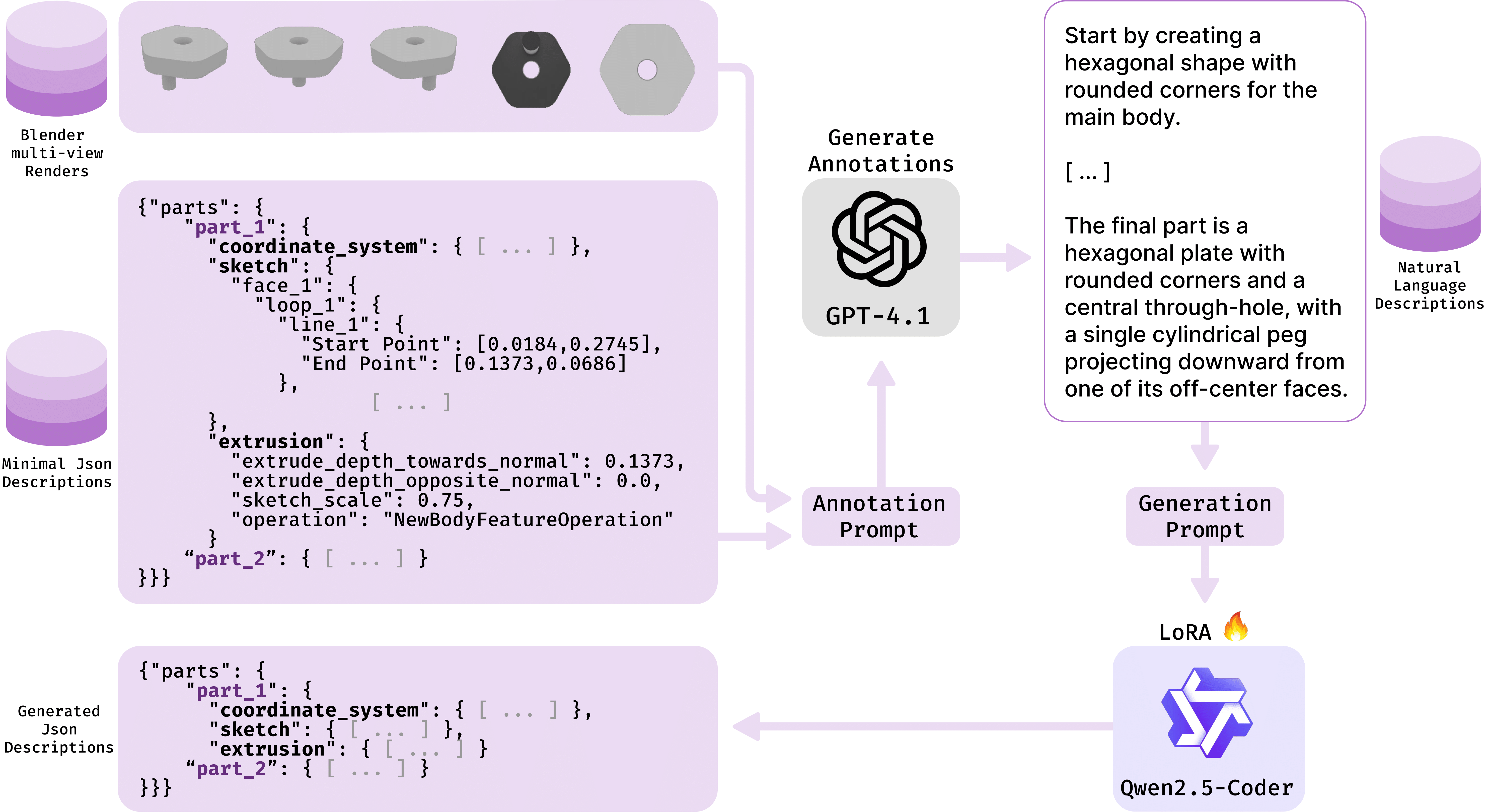
Reinforcement Learning for Crystal Structure Design
Currently exploring offline and online reinforcement learning methods for learning a policy for sequentially designing crystal structures. 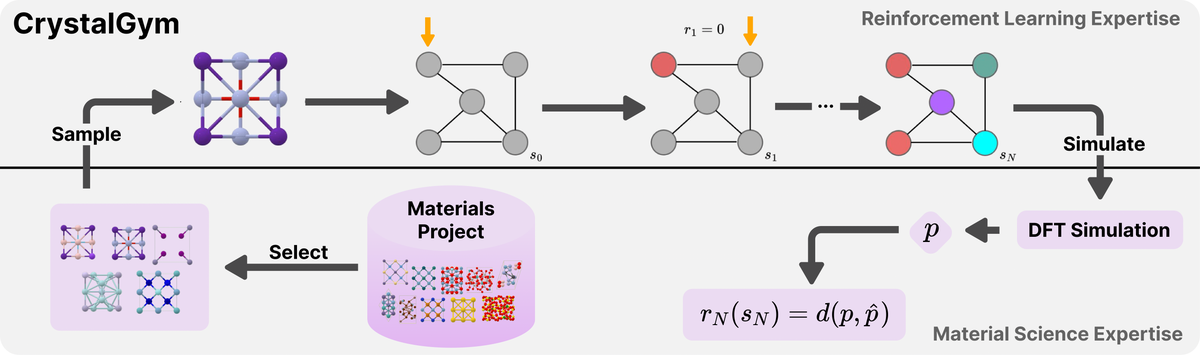
Graph generative models for binding site-specific molecule generation
Designing novel deep graph generative model for generating new ligand molecules that can bind to a given target receptor binding site. 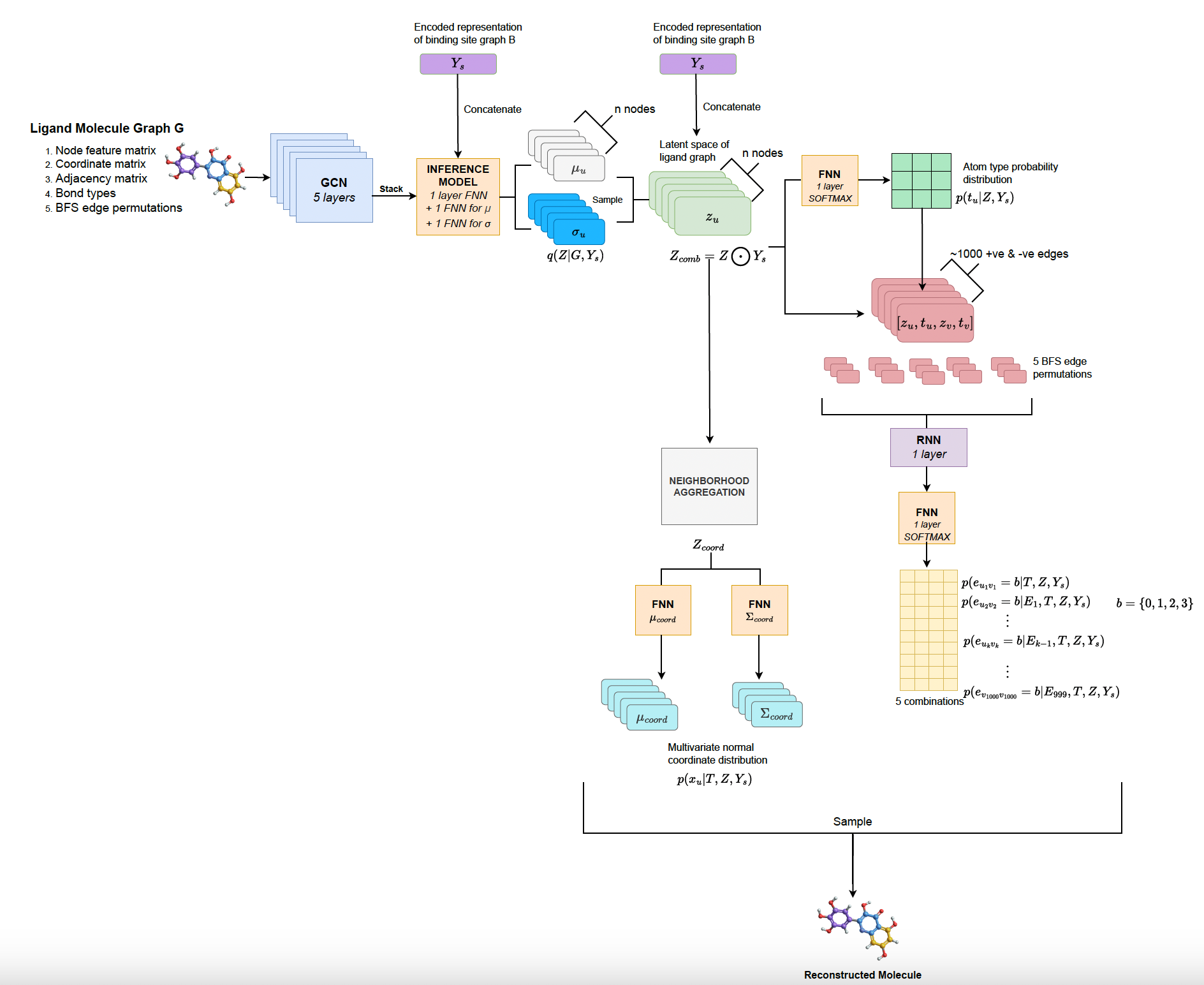
Analysis of drug response and gene expression data of AML cells
Computational methods to identify drug-gene correlations and molecules that can induce leukemic cell maturation. 
Incorporating Geometry into Score-Based Model for Crystal Structure Design
Two ways to incorporate crystal symmetry information as an inductive bias into a generative model for crystal structure design. 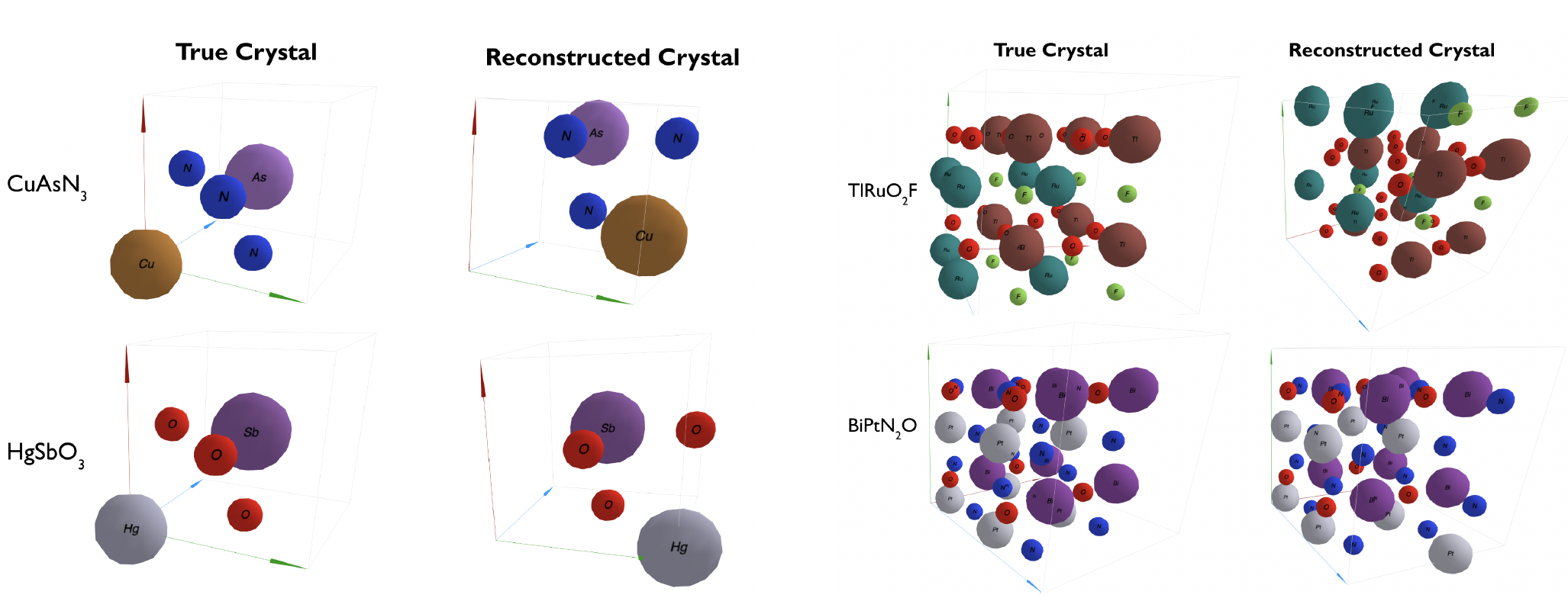
Analyzing the effects of visual representations in visual navigation tasks
Two ways for encoding visual inputs for navigation task using RL – pretraining a contrastive learning-based SimCLR model and VAE-based generative approach. 
Deep generative models for single-cell gene expression analysis
Evaluated state-of-the-art unsupervised deep learning techniques including variational autoencoders for single-cell gene expression data analysis. 
Generating drug-like molecules from gene expression signatures using transformer
Designed an attention-based transformer model for de novo generation of drug-like molecules that can induce a desired transcriptomic profile. Accepted as poster at MLCSB COSI, ISMB 2022. 
Parallel analyses of canonic polyadic tensor decomposition algorithm
CPU- and GPU-level parallelization of tensor decomposition algorithm using OpenMP and OpenACC. 
