Reinforcement Learning for Crystal Structure Design
Currently exploring offline and online reinforcement learning methods for learning a policy for sequentially designing crystal structures. 
Currently exploring offline and online reinforcement learning methods for learning a policy for sequentially designing crystal structures. 
Designing novel deep graph generative model for generating new ligand molecules that can bind to a given target receptor binding site.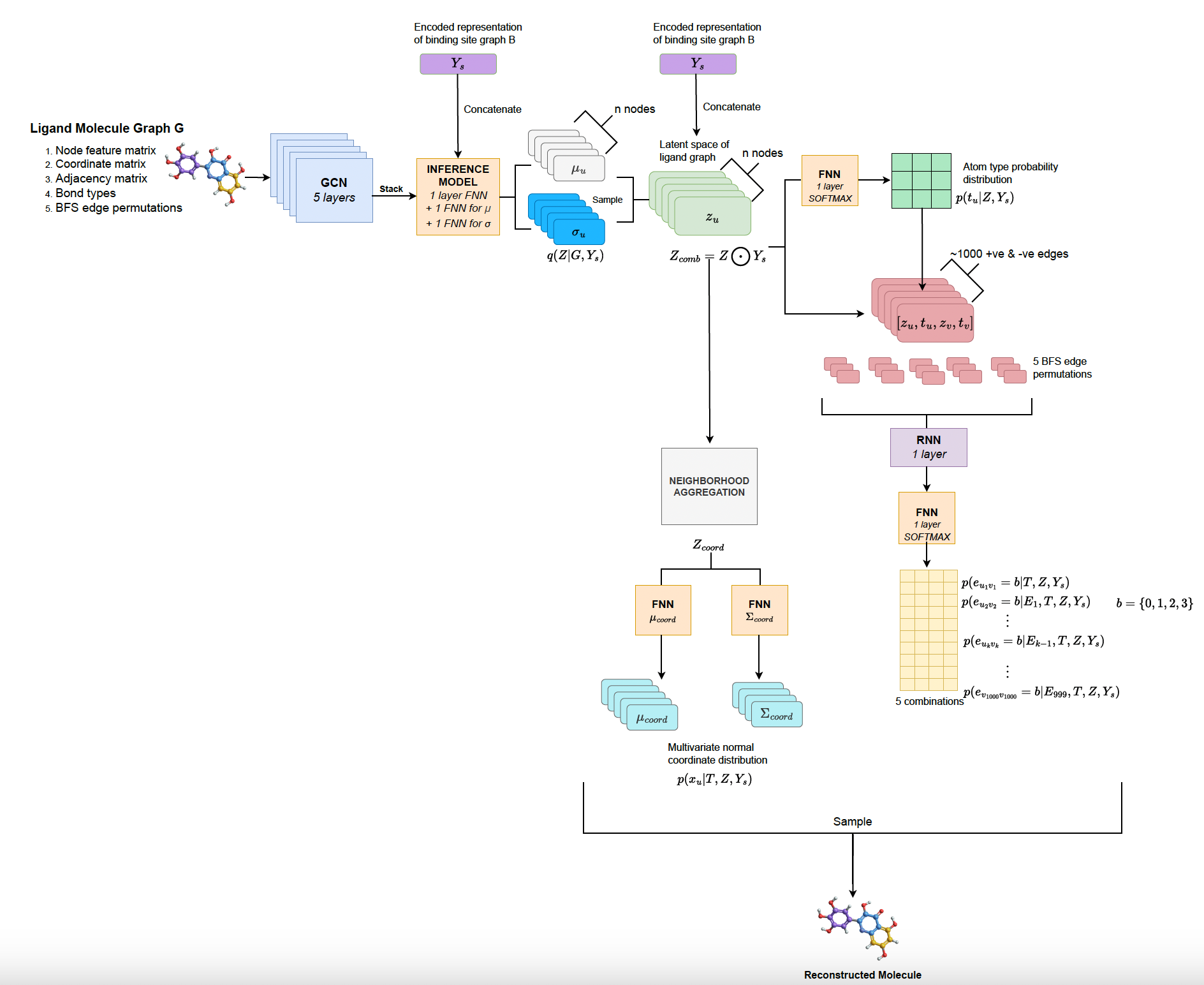
Computational methods to identify drug-gene correlations and molecules that can induce leukemic cell maturation.
Two ways to incorporate crystal symmetry information as an inductive bias into a generative model for crystal structure design.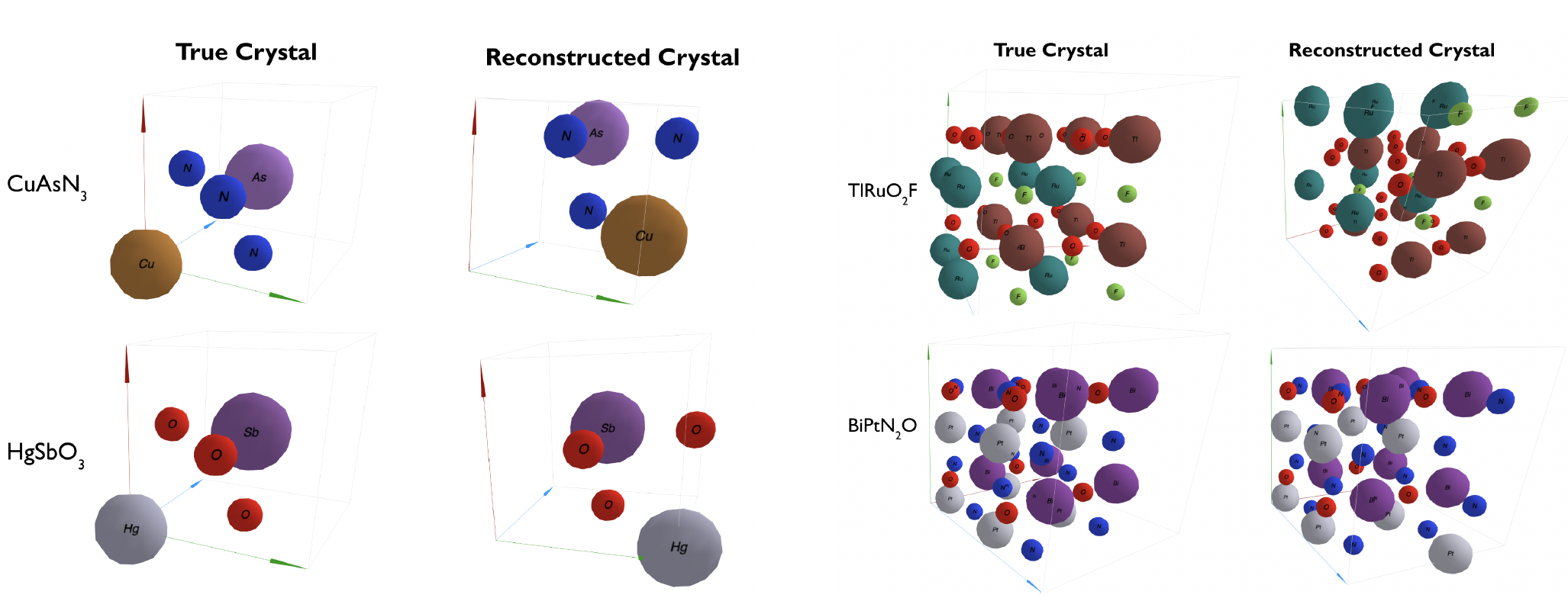
Two ways for encoding visual inputs for navigation task using RL – pretraining a contrastive learning-based SimCLR model and VAE-based generative approach.
Evaluated state-of-the-art unsupervised deep learning techniques including variational autoencoders for single-cell gene expression data analysis. 
Designed an attention-based transformer model for de novo generation of drug-like molecules that can induce a desired transcriptomic profile. Accepted as poster at MLCSB COSI, ISMB 2022. 
CPU- and GPU-level parallelization of tensor decomposition algorithm using OpenMP and OpenACC. 
Identified somatic mutations in RNA-sequencing data of human oral squamous cell carcinoma samples.